Polars, DuckDB, Pandas, Modin, Ponder, Fugue, Daft — which one is the best dataframe and SQL tool?
Comparing open-source dataframe and SQL frameworks for data engineering, machine learning and analytics
Tabular format with rows and columns, popularized by relational databases and Microsoft Excel, is an intuitive way of organizing and manipulating data for analytics.
There are two main ways of transforming and analyzing tabular data — SQL and dataframes (sorry, Excel!).
SQL is a declarative language for querying datasets of any size. It’s versatile, mature, time-tested, and widely adopted across various professions, organizations and tools.
However, some data transformations are more easily expressed using dataframes defined in an imperative language such as Python. This includes pivoting and melting (reverse process to pivoting), dealing with missing values or time series, visualizing and iteratively exploring data, or applying custom functions and machine learning algorithms. For such use cases, dataframes offer a great addition to SQL-based processes allowing in-memory computation and integrating well with data science workflows.
SQL vs. dataframes or SQL and dataframes?
In the past, SQL and dataframes were two separate worlds. SQL was used by data engineers for querying data lakes and data warehouses, while dataframes were used by ML engineers for in-memory computation and data science. Nowadays, this is changing. The lines between SQL and dataframes are becoming increasingly blurry.
🐼 While pandas is the most popular (“default”?) dataframe interface in Python, it can also execute SQL queries and write to SQL databases as long as you configure a SQLAlchemy engine or connection.
🐻 Polars is a lightning fast DataFrame library and in-memory query engine that allows you to query tabular data as dataframes in Python, Rust, Node.js, but it also provides a SQL context to run transformations in plain SQL.
🦆 DuckDB is an in-process OLAP database management system. While SQL is the “mother tongue” of MotherDuck and DuckDB, the tool seamlessly integrates with dataframe libraries such as pandas, Polars, Vaex, Ibis and Fugue, and provides an imperative Python client API on top of pandas DataFrames, Polars DataFrames and Arrow tables.
🎶 Fugue is a Python interface for distributed transformations over dataframes but it also can execute Fugue-SQL on top of local (pandas, Polars, DuckDB) or distributed (Spark, Dask, BigQuery, and more) dataframes.
💦 Modin provides a drop-in replacement for pandas, and their commercial offering (Ponder) can translate your pandas code to SQL under the hood and run it on DuckDB or a cloud data warehouse such as BigQuery or Snowflake.
✨ Spark SQL makes the distinction between dataframes and SQL almost invisible. To the end user, everything is a Spark dataframe regardless of whether you use SQL, Java, Scala, Python, or Pandas API.
❄️ Snowpark is Snowflake’s framework for building distributed dataframes. It allows writing custom imperative code in Python, Java, or Scala and pushes down computation to Snowflake as if you would write and execute SQL.
🔸 dbt is primarily a SQL-based data transformation tool but it also has a Python model abstraction to support… you guessed it, dataframes!
By now, you should be convinced that both SQL and dataframes can be used in tandem across a variety of data tools. But which one should you use? And when?
Let’s look at some of the tools mentioned above and explore the strengths of their table abstractions. To show the syntax of each framework, we’ll demonstrate how each of these tools can be used to find best-selling products. To do that, we need to follow these steps:
- Read multiple CSV files containing sales data
- Concatenate those into one table
- Join that table with products stored in a separate CSV file
- Find 10 best-selling products (in terms of order volume) by aggregating data
- Export the result in a JSON format so that it can be sent via an API call to operational systems (e.g., product recommender system).
You can download a zip file with data used in this article from the kestra-io/datasets repository.
Let’s get started with the most popular dataframe library in Python — pandas.
Pandas
The concept of dataframes has been initially established in the R programming language. Pandas, released in 2008, has brought dataframes to Python. Due to growing Python’s popularity, pandas has, over time, become the default choice for many data science and analytics tasks.
Despite the wide adoption, pandas has several drawbacks, which inspired many new open-source and commercial products. The creator of pandas alone, Wes McKinney, famously wrote “10 Things I Hate About pandas” on his blog, including aspects such as poor memory management, inefficient data types, and lack of query planning and multicore execution.
Here is our example use case built in Pandas:
import pandas as pdimport glob
csv_files = glob.glob("dataframes/2023*.csv")
dfs = [pd.read_csv(f) for f in csv_files]orders = pd.concat(dfs, axis=0, ignore_index=True)products = pd.read_csv("dataframes/products.csv")df = orders.merge(products, on="product_id", how="left")
top = ( df.groupby("product_name", as_index=False)["total"] .sum() .sort_values("total", ascending=False) .head(10))
top.to_json("bestsellers_pandas.json", orient="records")When writing pandas code, you need to be aware of the index. Pandas index has its pros and cons — just make sure to keep it in mind when writing Pandas code. For instance, without adding as_index=False, the result in this example would be a pandas Series rather than a DataFrame object, and saving the result to JSON would result in a list of values without matching product IDs.
What about SQL? You can execute SQL queries to load data from external databases using SQLAlchemy. However, pandas doesn’t provide a SQL-based interface to manipulate and query data — SQL is only limited to fetching data into a dataframe.
Polars
Polars is a blazingly fast DataFrame library written in Rust. Ritchie Vink, the creator of Polars, has improved upon pandas’ design choices and built a vectorized OLAP query engine that has excellent performance and memory usage.
The key drivers behind Polar’s performance (and popularity) are:
- The query optimizer that allows lazy evaluation so that users don’t need to worry about optimizations — Polars figures out the optimal path to run a query. According to Polars, without a query optimizer, other dataframe implementations ignore several decades of database research.
- Parallelism - Polars leverages lazy evaluation to parallelize query execution across all cores of your machine.
- Memory- and cache-efficient columnar format — pandas’ approach of storing arrays, strings, and structs as Python objects results in a single-threaded execution. In contrast, Polars uses data types based on Apache Arrow and can “put your machine to work”. By leveraging the streaming API, Polars can even work with larger-than-memory datasets.
Here is the representation of our use case in Polars:
import polars as plimport glob
csv_files = glob.glob("dataframes/2023*.csv")
df = ( ( pl.concat( [pl.scan_csv(f) for f in csv_files] ) .join( pl.scan_csv("dataframes/products.csv"), on="product_id", how="left" ) ) .groupby("product_name") .agg(pl.col("total").sum().alias("total")) .sort("total", descending=True) .limit(10)).collect()
df.write_json("bestsellers_polars.json", row_oriented=True)Polars API is composable and you don’t have to worry about the index (in fact, it doesn’t exist!). Writing Polars code feels as intuitive as the dplyr package in R.
The solution to our use case in Polars ran twice as fast as the one from pandas. This doesn’t matter as much in this toy example, but when dealing with large datasets, the difference becomes quite significant.
What about SQL? Polars provides SQL context supporting many operations. Here is the same example as before but using a combination of Python and SQL:
import polars as plimport glob
csv_files = glob.glob("dataframes/2023*.csv")
ctx = pl.SQLContext( orders=pl.concat([pl.scan_csv(f) for f in csv_files]), products=pl.scan_csv("dataframes/products.csv"), eager_execution=True,)
query = """SELECT product_name, sum(total) as totalFROM ordersLEFT JOIN products USING (product_id)GROUP BY product_nameORDER BY total DESCLIMIT 10"""
df = ctx.execute(query)df.write_json("bestsellers_polars.json", row_oriented=True)This example seamlessly combines Python and SQL code. You can see here that the differences between the “dataframe world” and the “SQL world” are disappearing. You can use both SQL and dataframes together — it’s no longer SQL OR dataframes, it’s now SQL AND dataframes.
What about distributed compute? Currently, the query engine can run only on a single machine. However, Polars has recently announced that they started a company. In the future, you can expect a distributed version of Polars available as a managed service.
DuckDB
DuckDB is an in-process OLAP DBMS designed to be embedded into applications. It is written in C++ and has client libraries for Python, SQL, Java, and even Swift (enabling DuckDB in mobile applications).
DuckDB SQL dialect supports complex types (arrays, structs), window functions, nested correlated subqueries, and many (many!) more. You can even execute SQL on top of your pandas or Polars dataframe.
Here is the same example using DuckDB in plain SQL:
COPY (SELECT product_name, sum(total) as total FROM '2023*.csv' LEFT JOIN 'products.csv' USING (product_id) GROUP BY product_name ORDER BY total DESC LIMIT 10)TO 'bestsellers_duckdb.json' (FORMAT JSON, ARRAY TRUE);This SQL code wins in terms of simplicity, readability, and conciseness. If you prefer Python over SQL, though, DuckDB supports that too:
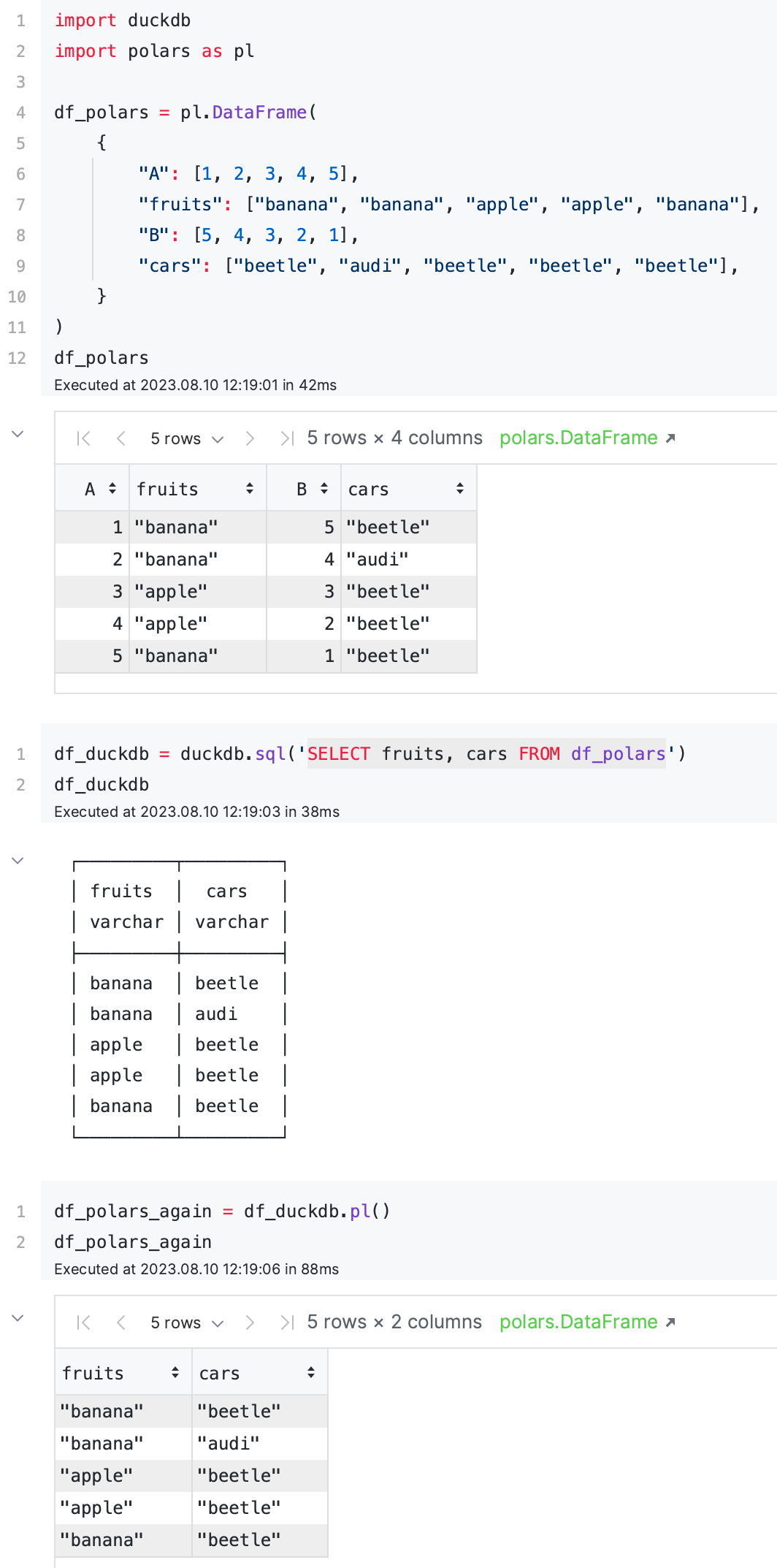
import duckdb
products = duckdb.read_csv("dataframes/products.csv")conn = duckdb.connect()orders = duckdb.sql("SELECT * FROM 'dataframes/2023*.csv'")
df = ( orders.join(products, condition="product_id", how="left") .aggregate("SUM(total) AS total, product_name", "product_name") .order("total DESC") .limit(10)).pl()
df.write_json("bestsellers_duckdb_py.json", row_oriented=True)Writing this “more Pythonic” code didn’t feel as intuitive to me as writing DuckDB SQL. I also had to convert the query result to a Polars (or pandas) DataFrame to export it as a JSON file.
Overall, DuckDB SQL is fantastic. It’s versatile and fast. But if you prefer to write idiomatic Python code without SQL, Polars might be a better fit for you. At the time of writing, it seems fair to compare the two as follows:
- for lightning fast single-node data transformations in SQL, use DuckDB
- for lightning fast single-node data transformations in Python, Node and Rust, use Polars
- for lightning fast single-node data transformations in general, use a combination of DuckDB and Polars at the same time.
As mentioned in the introduction, I believe that, over time, the differences here will continue to disappear. I can imagine that DuckDB will continue improving Python’s ergonomics, and Polars will extend its SQL support.
It’s also worth mentioning that DuckDB integrates exceptionally well with Polars — just add .pl() to convert your DuckDB table to a Polars DataFrame. You can switch between Polars and DuckDB with zero copy thanks to Apache Arrow. I see using both Polars and DuckDB as complementary, not competitive. Just look at this example from DuckDB docs:
What about distributed compute? DuckDB is an in-process single-node database. MotherDuck is a SaaS service that scales DuckDB to the cloud. Check our DuckDB vs. MotherDuck guide and our blueprints to learn more about various DuckDB use cases for scheduled ETL and event-driven workflows.
Speaking of scaling your dataframes, let’s cover Modin, Ponder, Fugue, and Daft next.
Modin
Modin is a drop-in replacement for pandas that scales to multiple cores and distributed clusters. It is built on top of Ray, Dask and DuckDB. It is a great way to scale pandas without having to rewrite your code.
Here is the same example as shown before with pandas but using Modin:
import modin.pandas as pdimport glob
csv_files = glob.glob("dataframes/2023*.csv")
dfs = [pd.read_csv(f) for f in csv_files]orders = pd.concat(dfs, axis=0, ignore_index=True)products = pd.read_csv("dataframes/products.csv")df = orders.merge(products, on="product_id", how="left")
top = ( df.groupby("product_id", as_index=False)["total"] .sum() .sort_values("total", ascending=False) .head(10))
top.to_json("bestsellers_modin.json", orient="records")We only had to change the import statement from import pandas as pd to import modin.pandas as pd to benefit from parallel compute on a locally initialized Ray cluster.
In this particular example, the Modin code was actually slower than pandas because the overhead of launching a Ray cluster was higher than the time it took to execute the query. However, if you work with large datasets and you already have a running cluster, Modin can deliver significant speedups to common pandas operations without having to rewrite any pandas code other than changing the import.
Let’s now look at Ponder, which is a commercial product built on top of Modin.
Ponder
The promise of Ponder is that you can execute your Modin code directly in your cloud data warehouse, such as BigQuery or Snowflake.
I was trying to use Ponder with BigQuery. To do that, I needed to sign up, authenticate my terminal with Ponder’s API token and install a bunch of Python libraries:
pip install ponderpip install google-cloud-bigquerypip install google-cloud-bigquery-storagepip install db_dtypesThen, the following code configures the BigQuery connection:
import ponder
ponder.init()
from google.cloud import bigqueryfrom google.cloud.bigquery import dbapifrom google.oauth2 import service_accountimport json
db_con = dbapi.Connection( bigquery.Client( credentials=service_account.Credentials.from_service_account_info( json.loads(open("/Users/anna/dev/gcp/credentials.json").read()), scopes=["https://www.googleapis.com/auth/bigquery"], ) ))
ponder.configure(default_connection=db_con, bigquery_dataset="dataframes")Finally, I was able to run the code for our use case — exactly the same Modin code as shown in the last section:
import modin.pandas as pdimport glob
csv_files = glob.glob("dataframes/2023*.csv")
dfs = [pd.read_csv(f) for f in csv_files]orders = pd.concat(dfs, axis=0, ignore_index=True)products = pd.read_csv("dataframes/products.csv")df = orders.merge(products, on="product_id", how="left")
top = ( df.groupby("product_id", as_index=False)["total"] .sum() .sort_values("total", ascending=False) .head(10))
top.to_json("bestsellers_ponder.json", orient="records")It took some time but eventually Ponder was able to finish the task. However, it also created 14 intermediate tables in my BigQuery dataset (yes, 14!) to perform that simple task. See the screenshot below for reference.
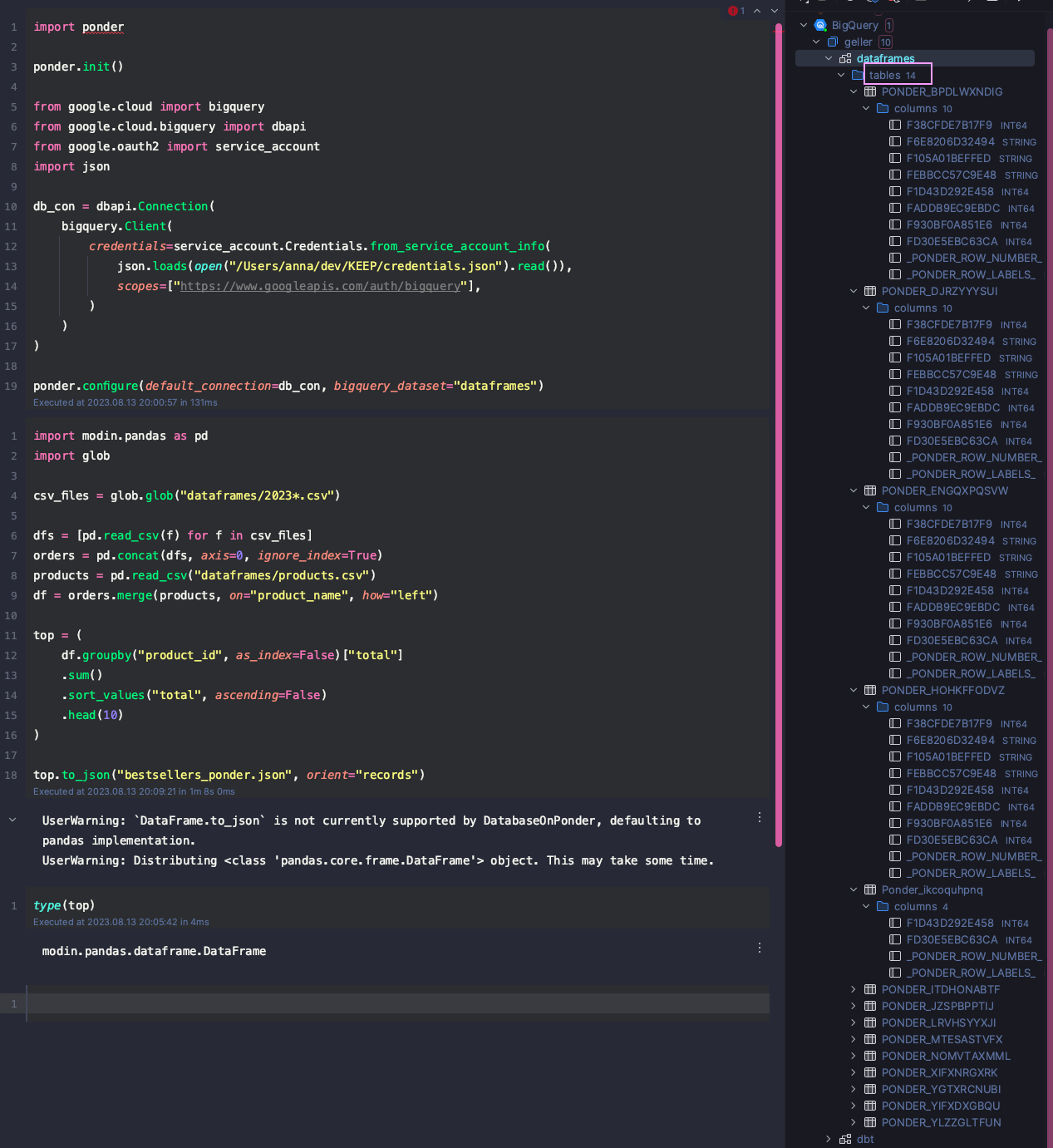
My overall impression is that Ponder seems like an interesting product for big data as it executes everything in a cloud data warehouse such as BigQuery. However, for our use case, Ponder felt a bit too heavy.
Next, let’s look at Fugue, which also provides a distributed dataframe abstraction that can run on top of BigQuery, Dask, Spark, DuckDB, and more.
Fugue
Fugue is described as a unified interface for distributed computing. You can execute Python, pandas, and SQL code on Spark and Dask with minimal rewrites.
Similarly to Modin, Fugue can help you scale pandas’ code across distributed compute clusters. In contrast to Modin, though, Fugue is not a drop-in replacement for pandas. Fugue’s core contributors believe that pandas-like interfaces are sub-optimal for distributed computing.
This means that you can continue writing pandas code, but Fugue will translate it at runtime to Spark or Dask syntax in order to distribute the underlying computations.
Fugue also encourages a mix of Python and SQL code by providing Fugue SQL on top of the core dataframe abstraction.
You can install Fugue and extensions for the specific distributed compute engine as follows:
pip install fuguepip install "fugue[dask]"Let’s now try to solve our use case with Fugue on Dask:
import fugue.api as faimport glob
csv_files = glob.glob("dataframes/2023*.csv")
def run(engine=None): with fa.engine_context(engine): orders = fa.load(csv_files, header=True) products = fa.load("dataframes/products.csv", header=True) df = fa.join(orders, products, how="left_outer", on=["product_id"]) res = fa.fugue_sql( """ SELECT product_name, SUM(total) as total FROM df GROUP BY product_name ORDER BY total LIMIT 10 """ ) fa.save(res, "bestsellers_fugue.json")
run(engine="dask") # runs on DaskThe biggest strength of Fugue is that switching between pandas, Spark, Dask, and many other compute engines is as simple as changing the engine variable:
run(engine="spark") # runs on Sparkrun(engine="duckdb") # runs on DuckDBrun() # runs on pandasMany Fugue users also appreciate the ability to interleave SQL and Python code within the same data transformation. However, if you prefer pure SQL, Fugure supports that too. Here is the same example using only SQL:
import fugue.api as fa
def run(engine=None): query = """ orders = LOAD "dataframes/2023*.csv" (header=true) products = LOAD "dataframes/products.csv" (header=true)
df = SELECT product_name, CAST(total AS INT) AS total FROM orders LEFT JOIN products ON orders.product_id = products.product_id
res = SELECT product_name, SUM(total) AS total FROM df GROUP BY product_name ORDER BY total LIMIT 10
SAVE TO "bestsellers_fugue.json" """
fa.fugue_sql(query, engine=engine)
run(engine="duckdb")Daft
Daft is another recently started open-source DataFrame project. At first glance, Daft seems to be as powerful and expressive as Polars while also being able to support distributed computation as Spark, Dask, and Ray.
To get started, you can install it using:
pip install getdaftpip install charset_normalizer --upgrade # I had to add this on M1Initially, I was getting exit code 132 even when running a simple script only reading a tiny CSV file. It turned out that this was an M1-mac issue running Python (installed with brew) in emulation mode with Rosetta (kudos to Jay Chia for help with that!). Daft is running a bunch of optimizations that break in emulation mode, hence the initial error.
After installing Conda for M1/M2, running python -c "import sysconfig; print(sysconfig.get_platform())" validated that I was now on the correct Python distribution for my architecture: macosx-11.1-arm64.
Here is the Daft syntax for our use case:
import daft
df = ( daft.read_csv("dataframes/2023_*.csv") .join(daft.read_csv("dataframes/products.csv"), on="product_id", how="inner") .groupby("product_name") .sum("total") .sort("total", desc=True) .limit(10))df.to_pandas().to_json("bestsellers_daft.json", orient="records")print(df.collect())There is no DataFrame.write_json() method yet, so you need to convert Daft DataFrame to pandas to get the result in a JSON format.
Overall, the project seems to be heading in an interesting direction. The table below shows how Daft positions itself among other dataframe libraries:
| Dataframe | Query Optimizer | Complex Types | Distributed | Arrow Backed | Vectorized Execution Engine | Out-of-core |
|---|---|---|---|---|---|---|
| Daft | Yes | Yes | Yes | Yes | Yes | Yes |
Pandas | No | Python object | No | optional >= 2.0 | Some(Numpy) | No |
Polars | Yes | Python object | No | Yes | Yes | Yes |
Modin | Eagar | Python object | Yes | No | Some(Pandas) | Yes |
Pyspark | Yes | No | Yes | Pandas UDF/IO | Pandas UDF | Yes |
Dask DF | No | Python object | Yes | No | Some(Pandas) | Yes |
Honorable mentions
We’ve covered a lot already. Here are some additional tools that support Dataframe and SQL workflows:
- Vaex - out-of-core hybrid Apache Arrow/NumPy DataFrame library for Python, ML, visualization and exploration of big tabular data at a billion rows per second.
- Spark SQL - a SQL interface to Spark dataframes. This is the most mature interface when it comes to seamlessly combining SQL with imperative code written in Python, Scala and Java. Spark was also the first dataframe framework that added SQL support.
- Pandas on Spark — while Spark SQL is mature, the Pandas API for Spark is somewhat limited with only around 60% coverage of what you can do with Pandas. Still, if you need some Pandas functionality while mainly using PySpark, it’s worth checking out. You can notice here that Spark is the most comprehensive among all these dataframe frameworks, covering Pandas API, DataFrame libraries in multiple lanuages, while also providing a reliable SQL interface on top of them.
- Dask and Ray — distributed frameworks to parallelize Python and ML applications. The dask-sql project provides a distributed query engine in Python, allowing a mixture of SQL operations and Python code. Ray supports SQL via an external open-source contribution called ray-sql. However, judging by the number of downloads, both
dask-sqlandray-sqlare not widely used yet. - R’s data.table — provides a high-performance version of base R’s
data.frametable abstraction. - bodo.ai is a SQL and Python data processing platform powered by MPI parallelization technologies. Initially, they promised faster performance than pandas simply by wrapping your functions with a
@bodo.jitPython decorator, but since then, they seemed to have largely switched their focus to SQL-based workflows with their Bodo SQL context. - Ibis — a Python framework that strives to offer the flexibility of Python analytics with the scale and performance of modern SQL. It’s neither a pandas-like dataframe nor SQL, but it gives you a way to write SQL queries in Python.
- Rapids - GPU-accelerated dataframes.
How should you choose among so many options?
Here is what you should consider when choosing a dataframe framework:
- What skills do people in your organization have? Are they more comfortable with SQL or Python?
- How much data do you have? Do you need to scale to multiple machines?
- What is the maturity of the framework? Is it actively maintained?
It’s always best to start simple. You can start with:
With these tools, you can easily transition to pandas and other frameworks when you need to, thanks to the Apache Arrow format.
If maturity is important to you, it’s still worth considering Spark. Even though the landscape is progressing quickly, Spark is still the most mature and comprehensive dataframe framework.
Using SQL and dataframes in your data pipelines
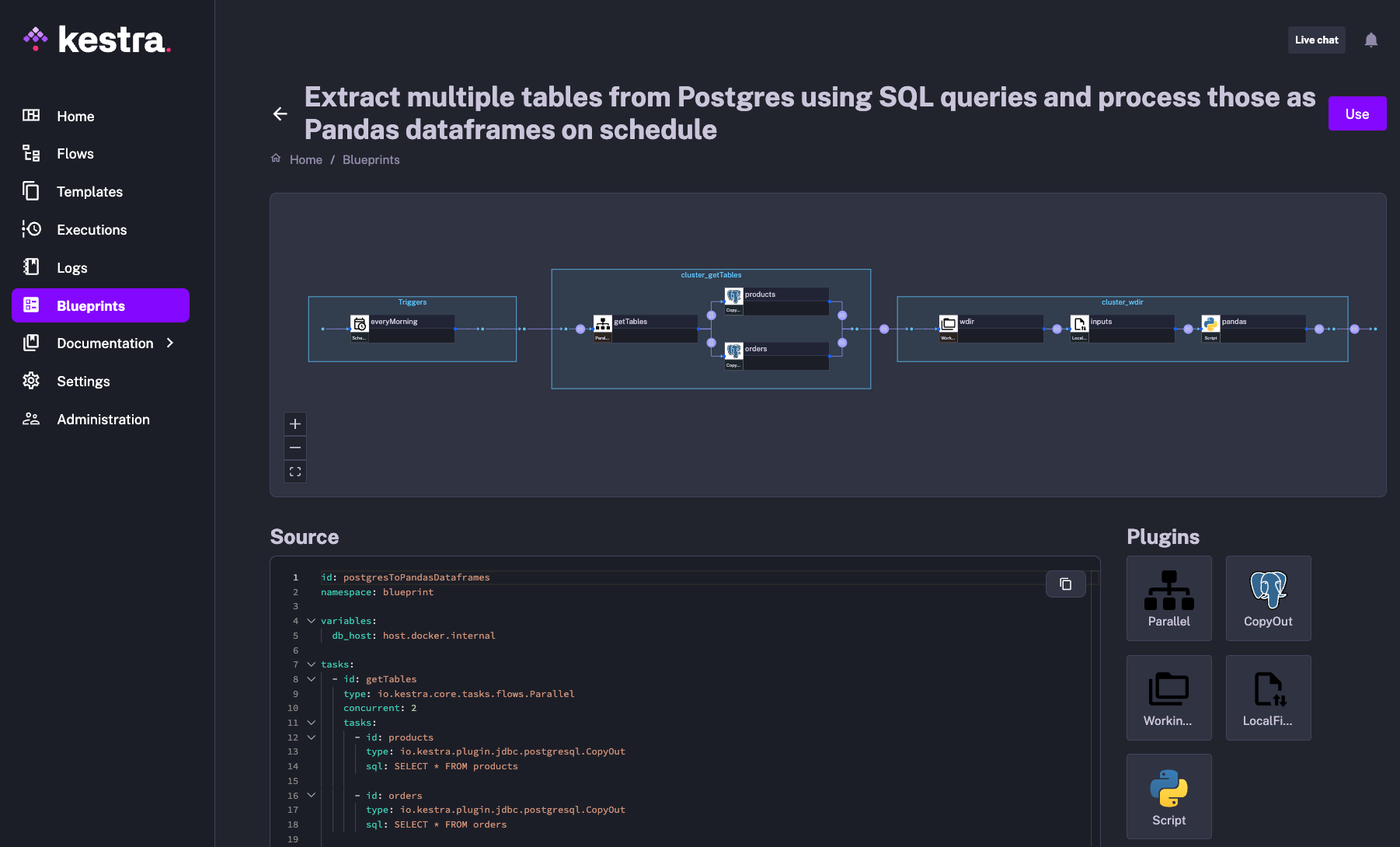
Kestra is a simple and fast open-source data orchestrator that can help you integrate your SQL and dataframe transformations into end-to-end data pipelines. The UI ships with a variety of blueprints that you can use to get started quickly.
The following blueprint shows how you can easily orchestrate your SQL and Pandas data transformations.
Summary
This post covered several open-source projects that support dataframes and SQL workflows. Which table abstraction is your favorite? Let us know in the community Slack. Make sure to check out Kestra’s blueprints to discover how to orchestrate both SQL and dataframe workflows. If you like Kestra, give us a star on GitHub and join the community.