Apache Iceberg Crash Course for AWS users: Amazon S3, Athena & AWS Glue ❤️ Iceberg

This crash course will guide you on how to get started with Apache Iceberg on AWS. By the end of this tutorial, you’ll be able to create Iceberg tables, insert and modify data stored in S3 in Parquet format, query data and table metadata in plain SQL, and declaratively manage the data ingestion process. Let’s get started.
What is Iceberg
Iceberg is an open table format. It’s a metadata layer that brings reliable transactions, schema evolution, version history, and data management to files in a data lake at a petabyte scale. You can think of Iceberg as an API between storage (e.g., parquet files on S3) and compute (Spark, Flink, DuckDB, Trino, Presto, Amazon Athena, Hive, BigQuery, Snowflake warehouse, etc.). You can define your table structure once on the API layer and then simultaneously query that table from anywhere (Snowflake, Spark, DuckDB, and more) without having to think about where data is physically located or how it is partitioned.
It still may sound too abstract, so let’s see it in action.
Iceberg demo with Amazon Athena and S3 from the AWS console
To follow along with this demo, you need an AWS account. We’ll be using Amazon Athena, Amazon S3, and the AWS Glue catalog.
If you don’t have an S3 bucket yet, you can create one following this AWS documentation.
Then, go to the Athena console and click “Edit Settings” to configure your query result location. This is required by Athena to e.g. cache query results. You can use the same bucket you created earlier as a location for query results.
To keep things simple, choose the same AWS region for both Athena and your S3 bucket.
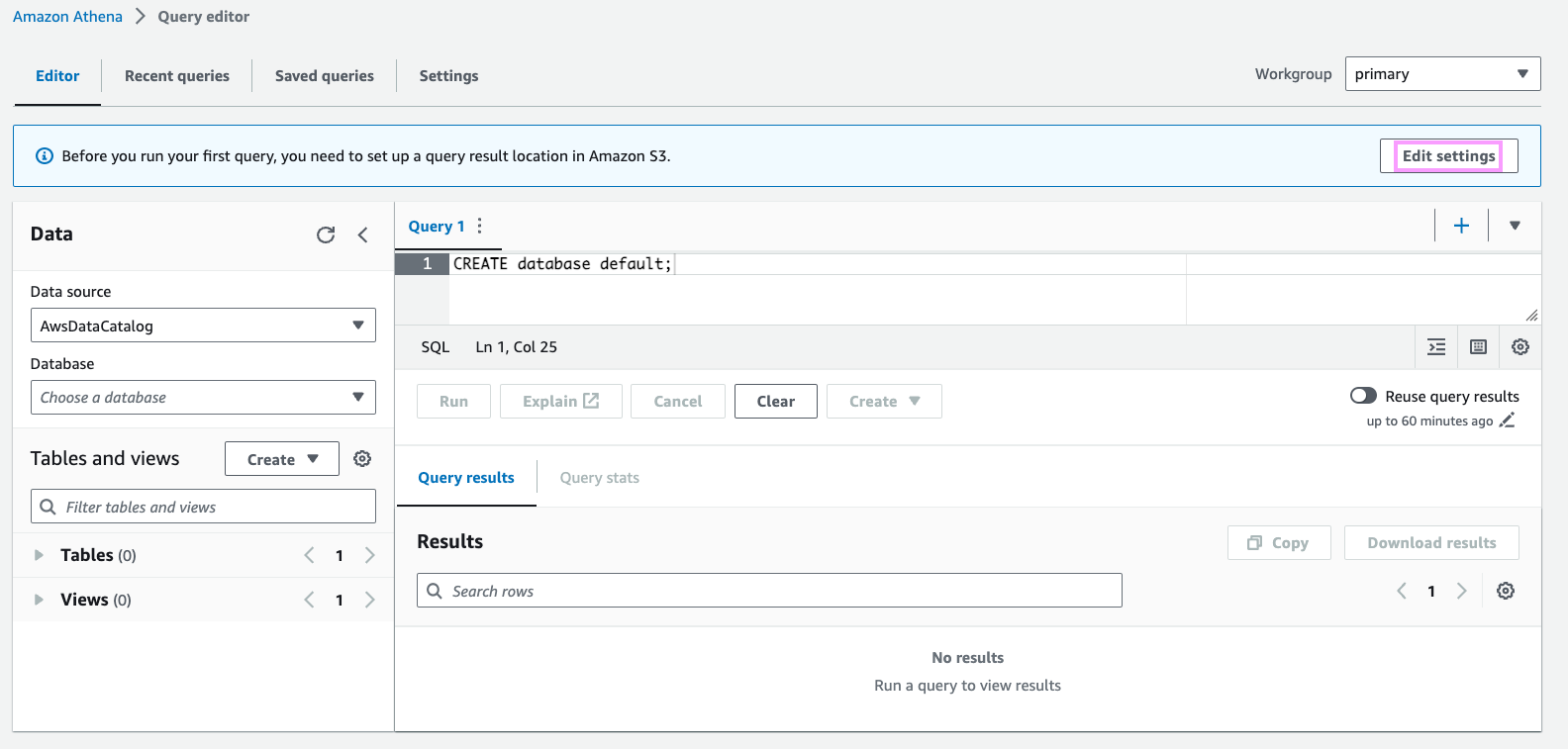
Create an Iceberg table
On the left side of the Athena query editor, you can select a database. Select the default database and run the following query:
CREATE TABLE fruits ( id bigint, fruit string, berry boolean, update_timestamp timestamp) PARTITIONED BY (berry)LOCATION 's3://kestraio/fruits/' -- adjust to your S3 bucket nameTBLPROPERTIES ('table_type'='ICEBERG');Let’s explain what the query does. First, we define the columns and their data types. The berry column is a boolean property that we’ll use for partitioning. It will divide the data into two partitions: berry=true and berry=false.
The LOCATION keyword determines where Iceberg will store data and metadata files:
- Data files contain the actual data stored in a
parquetformat - Metadata files contain metadata information stored in
jsonandavrofile formats.
The TBLPROPERTIES keyword tells Athena that we want to use the Iceberg format as a table type.
After executing this query, you should see a new Iceberg table called fruits in the Athena table list on the left.
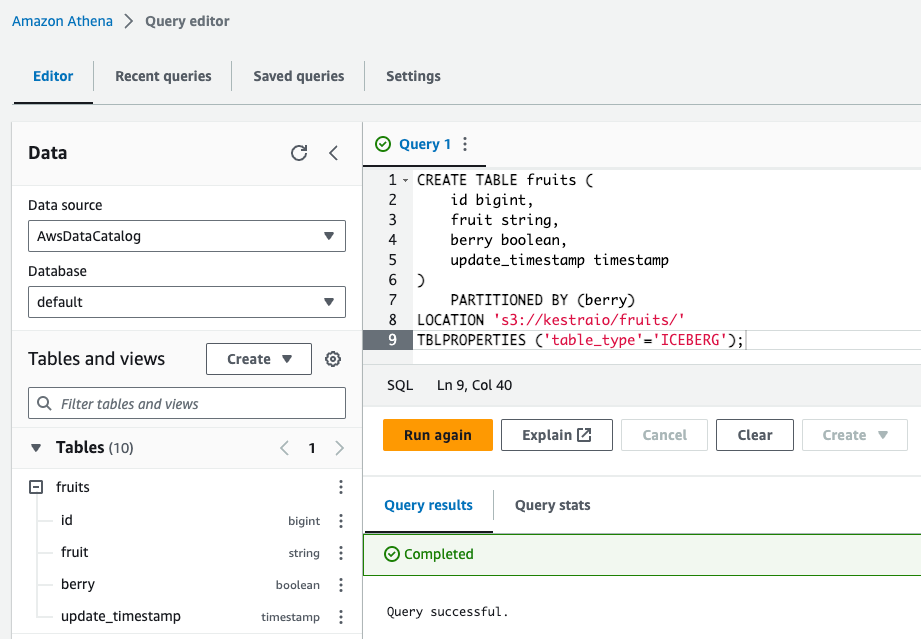
Insert new rows using INSERT INTO
So far, we’ve created an empty table. Let’s insert some data into it.
INSERT INTO fruits (id, fruit, berry) VALUES (1,'Apple', false), (2, 'Banana', false), (3, 'Orange', false), (4, 'Blueberry', true), (5, 'Raspberry', true), (6, 'Pear', false);Side note: botanically speaking, a banana is a berry. We’ll ignore that fact in the data to avoid confusion. 🍌
Let’s add a couple of new rows in a separate SQL statement to demonstrate later how Iceberg handles transactions and file partitioning.
INSERT INTO fruits (id, fruit, berry) VALUES (7,'Mango', false), (8, 'Strawberry', true), (9, 'Kiwi', false), (10, 'Cranberry', true);Finally, let’s inspect the data we’ve just inserted.
SELECT *FROM fruitsORDER BY berry;You should see the following results:
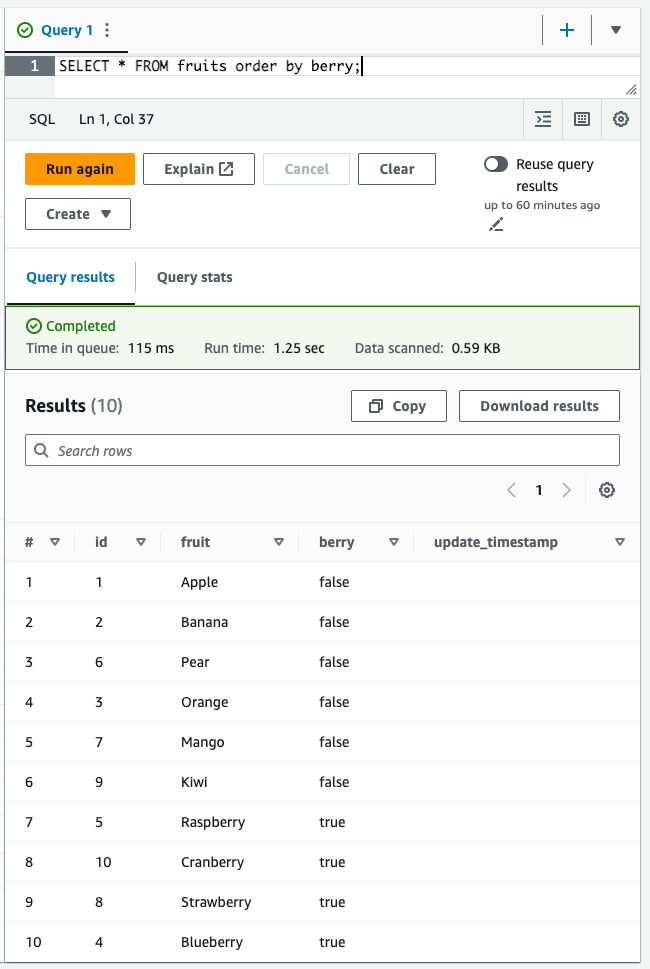
Marvellous! We were able to just insert rows to files in a data lake as if we were using a regular relational database. This is the power of Iceberg — it brings transactions to the data lake.
Modify rows using UPDATE and DELETE
Similarly to how Iceberg allowed us to perform row-level inserts, we can also modify data in the table using UPDATE and DELETE SQL statements.
Let’s imagine that, as Erik, you grew up eating Bilberries, thinking that those are just regular Blueberries.

Let’s correct that in our data:
UPDATE fruitsSET fruit = 'Bilberry'WHERE fruit = 'Blueberry';Let’s also remove the banana to avoid debates about whether a banana is actually a berry or not.
DELETE FROM fruitsWHERE fruit = 'Banana';Let’s validate what we have so far in the table:
SELECT *FROM fruitsORDER BY berry;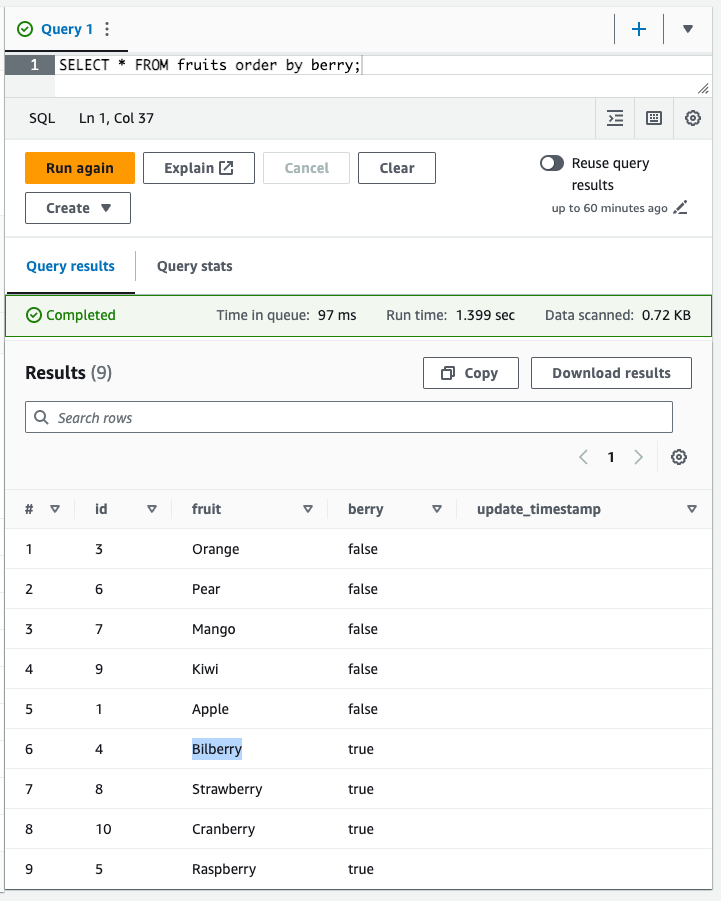
Excellent! We’ve just updated and deleted rows in our data lake as if it were a relational data store. Let’s now inspect the table metadata to see how Iceberg handles these operations under the hood.
Iceberg metadata
First, let’s look at files that are stored in our S3 bucket. You can explore them in the AWS console or using the AWS CLI.
aws s3 ls s3://yourbucket/fruits/ --recursive --summarize --human-readableYou should see 28 files structured in the following way:
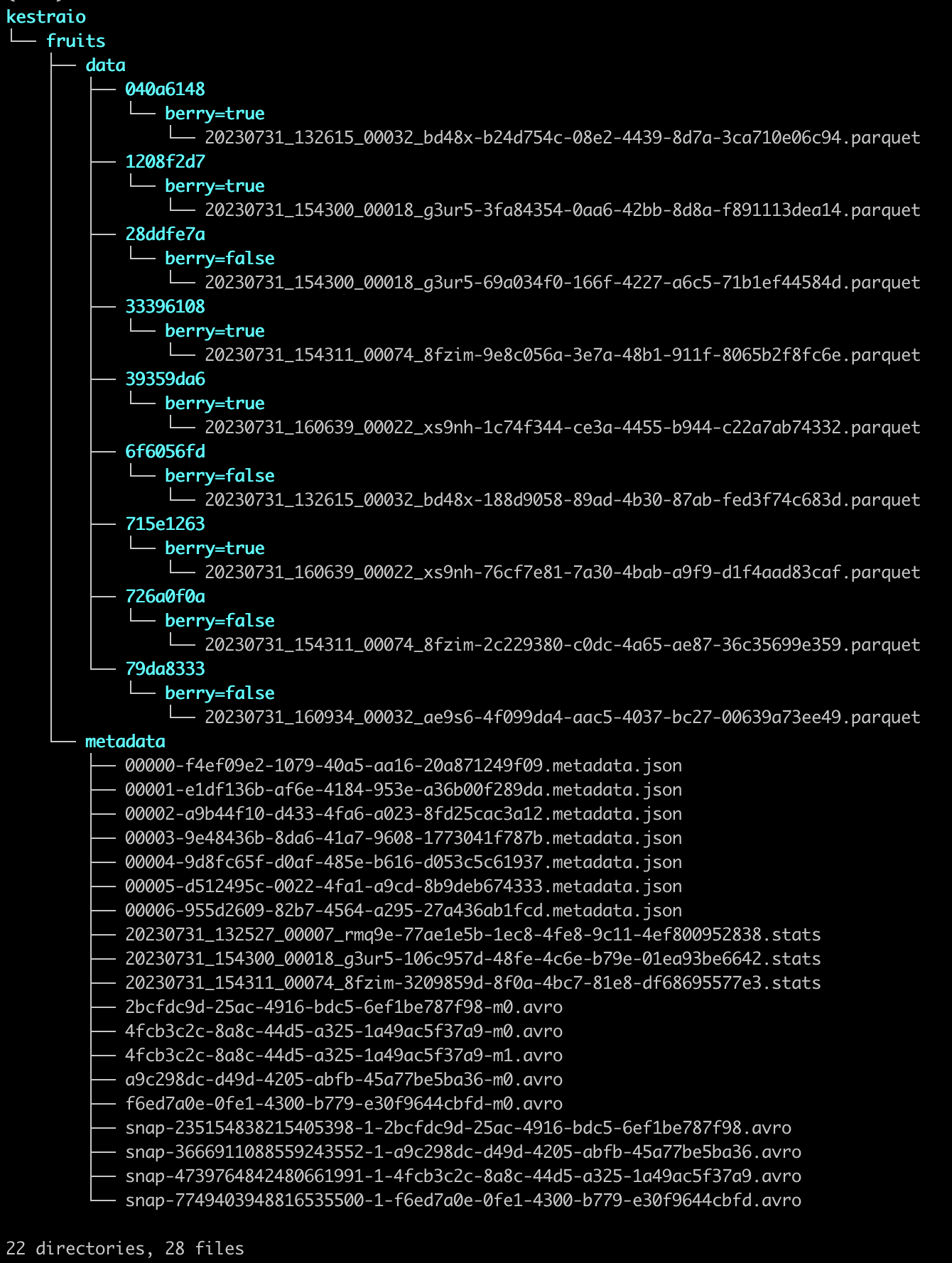
The data folder contains parquet files with the actual data, and the metadata folder contains Iceberg-internal metadata information.
Note that as an end-user, you shouldn’t have to worry about the storage layer. Those files are managed by Iceberg. Instead, you can use dedicated metadata properties that Iceberg exposes on each table, as shown in the code block below. This allows you to inspect the table’s history, partitions, snapshots, files on S3, and more — you can query all that metadata in plain SQL.
SELECT * FROM "fruits$files";SELECT * FROM "fruits$manifests";SELECT * FROM "fruits$history";SELECT * FROM "fruits$partitions";SELECT * FROM "fruits$snapshots";SELECT * FROM "fruits$refs";Let’s look at the table history:
SELECT *FROM "fruits$history";You should see four rows, which reflect two INSERTS, one UPDATE and one DELETE operations that we have executed so far. The fruits$snapshots provides a summary of each operation (showing which files or partitions have been added, modified or deleted), as you can see in the image below:
SELECT snapshot_id, summaryFROM "fruits$snapshots";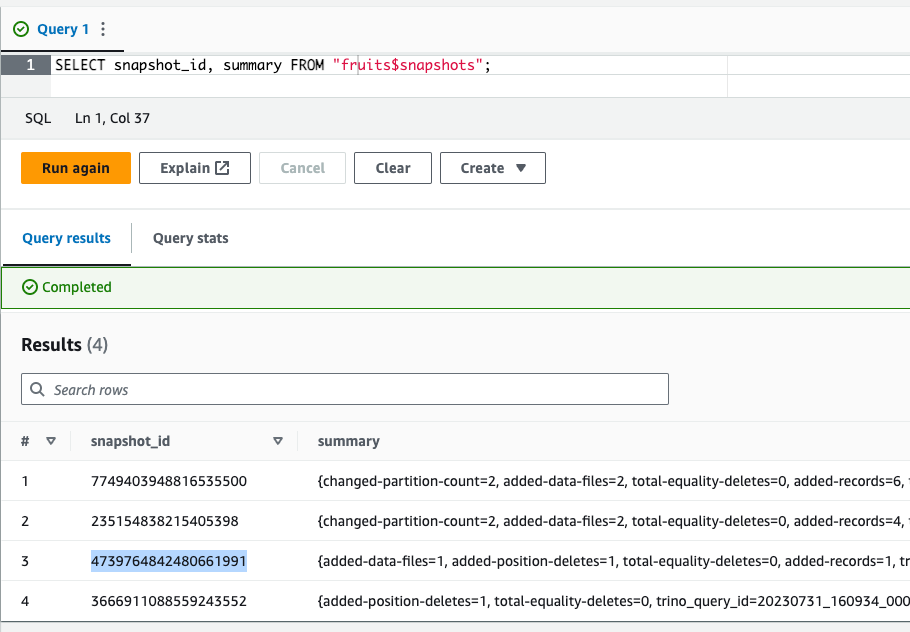
Let’s copy that snapshot ID and inspect the table as of the time when that snapshot was taken:
SELECT *FROM fruitsFOR VERSION AS OF 4739764842480661991;You can see that back then, Banana was still in our data:
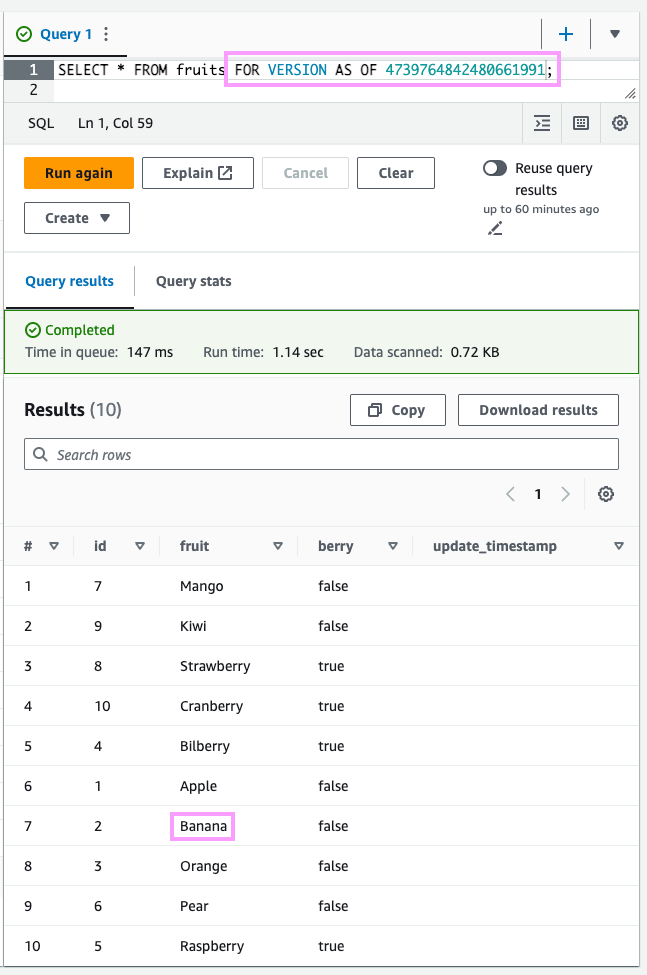
Time travel
Apart from inspecting a specific table’s snapshot version, you can also query the table as of a specific timestamp, effectively giving your data the ability to time-travel. How cool is that!
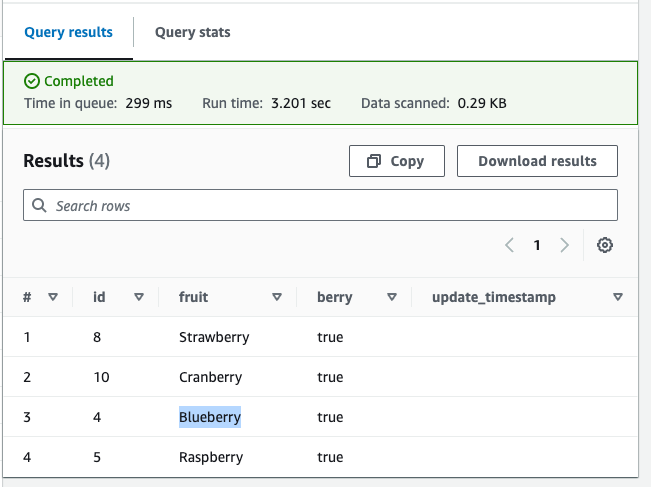
Let’s see what berries existed in our data 5 minutes ago:
SELECT *FROM fruitsFOR TIMESTAMP AS OF (current_timestamp - interval '5' minute)WHERE berry = true;Back then, we were still under the illusion that we grew up eating Blueberries, while these were actually Bilberries:
Bulk data ingestion
So far, we’ve been inserting data into our table row by row. However, data lakes are typically used for big data processed via batch or streaming jobs.
There are two common patterns of ingesting data to existing Iceberg tables: inserts and upserts. Both of them require loading data into a different (temporary) table before being ingested to the final destination via a separate query.
Let’s ingest new rows from a file into a temporary table. We’ll use pandas and awswrangler libraries to read data from a CSV file into a dataframe. Then, we’ll insert that dataframe into a temporary table called raw_fruits:
import awswrangler as wrimport pandas as pd
bucket_name = "kestraio"glue_database = "default"glue_table = "raw_fruits"path = f"s3://{bucket_name}/{glue_table}/"temp_path = f"s3://{bucket_name}/{glue_table}_tmp/"
wr.catalog.delete_table_if_exists(database=glue_database, table=glue_table)
df = pd.read_csv("https://huggingface.co/datasets/kestra/datasets/raw/main/csv/fruit.csv")df = df[~df["fruit"].isin(["Blueberry", "Banana"])]df = df.drop_duplicates(subset=["fruit"], ignore_index=True, keep="first")
wr.athena.to_iceberg( df=df, database=glue_database, table=glue_table, table_location=path, temp_path=temp_path, partition_cols=["berry"],)Don’t forget to adjust the S3 bucket name. Also, make sure to install the awswrangler library first (awswrangler already includes pandas and boto3):
pip install awswranglerWe can validate that the table was successfully created by running the following query:
SELECT *FROM raw_fruits;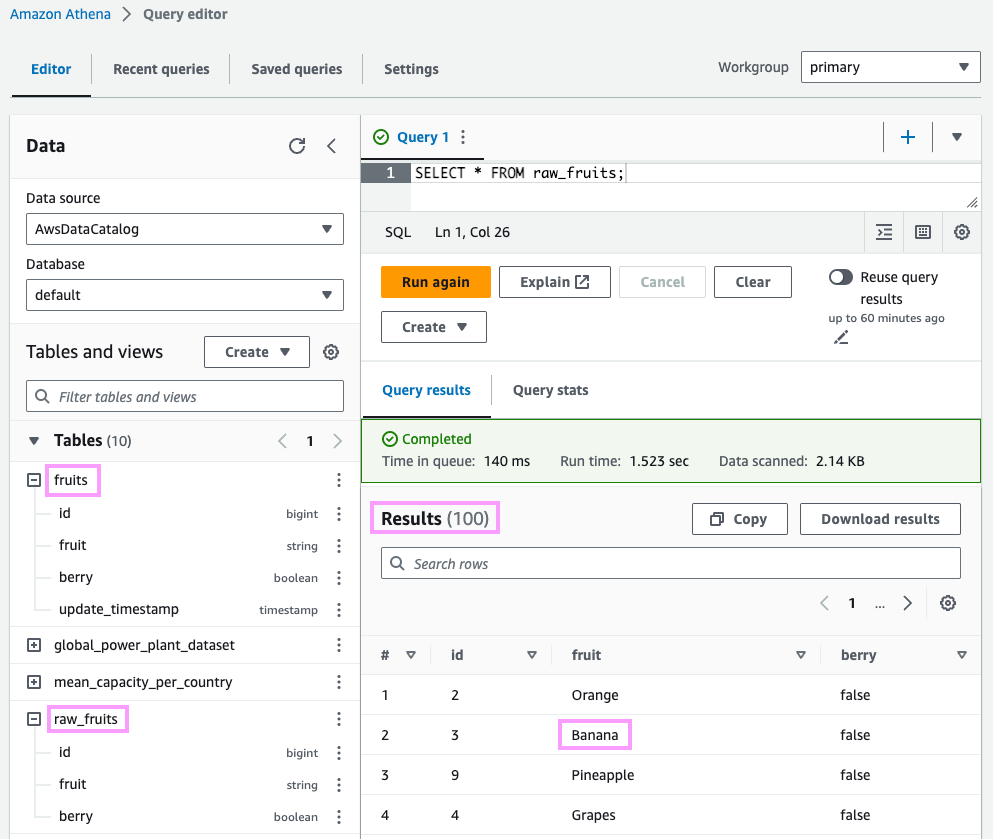
The data looks good. Alas, the banana strikes back! 🥊🍌 We’ll take care of that when building an end-to-end data pipeline in the final section.
Inserts and upserts in Iceberg
Here is how inserts and upserts work in Iceberg tables:
INSERT INTO fruits as SELECT * FROM raw_fruits— this insert pattern is useful when you want to simply append data to an existing table.MERGE INTO fruits as USING raw_fruits ON fruits.id = raw_fruits.id WHEN MATCHED THEN UPDATE SET * WHEN NOT MATCHED THEN INSERT *— this upsert pattern is useful when you want to update existing rows; for instance, if a specific fruit ID is now called differently (or is now considered a berry 🫐), you may want to update an existing entry rather than inserting it as a duplicate row.
Below, we use the MERGE pattern by merging the tables on the fruit name.
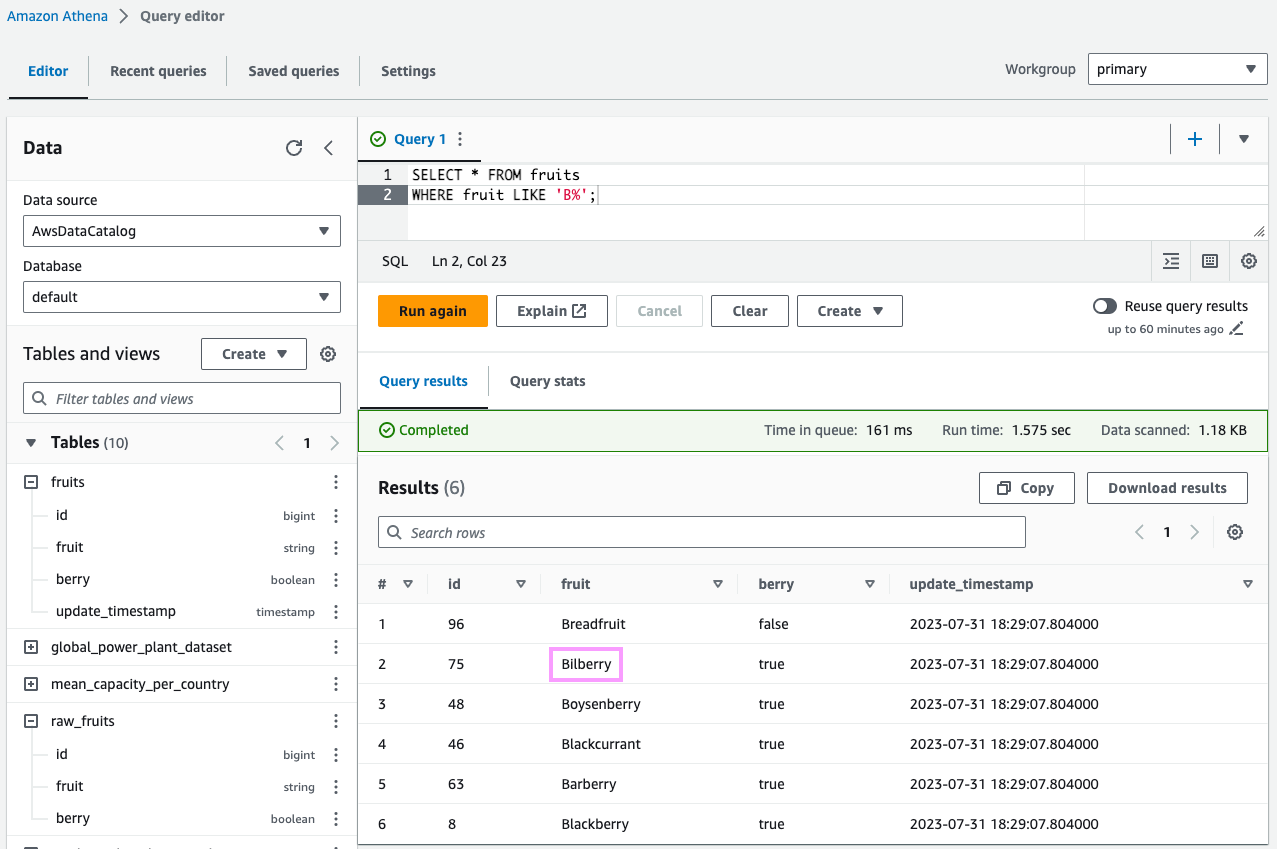
MERGE INTO fruits f USING raw_fruits r ON f.fruit = r.fruit WHEN MATCHED THEN UPDATE SET id = r.id, berry = r.berry, update_timestamp = current_timestamp WHEN NOT MATCHED THEN INSERT (id, fruit, berry, update_timestamp) VALUES(r.id, r.fruit, r.berry, current_timestamp);Let’s inspect all fruits that start with B (to check on our beloved Bilberry, Blueberry and Banana):
SELECT *FROM fruitsWHERE fruit LIKE 'B%';This looks great! We inserted new data and ensured that no Blueberries or Bananas made it to the final dataset.
Let’s look at the files generated so far by Iceberg:
SELECT *FROM "fruits$files";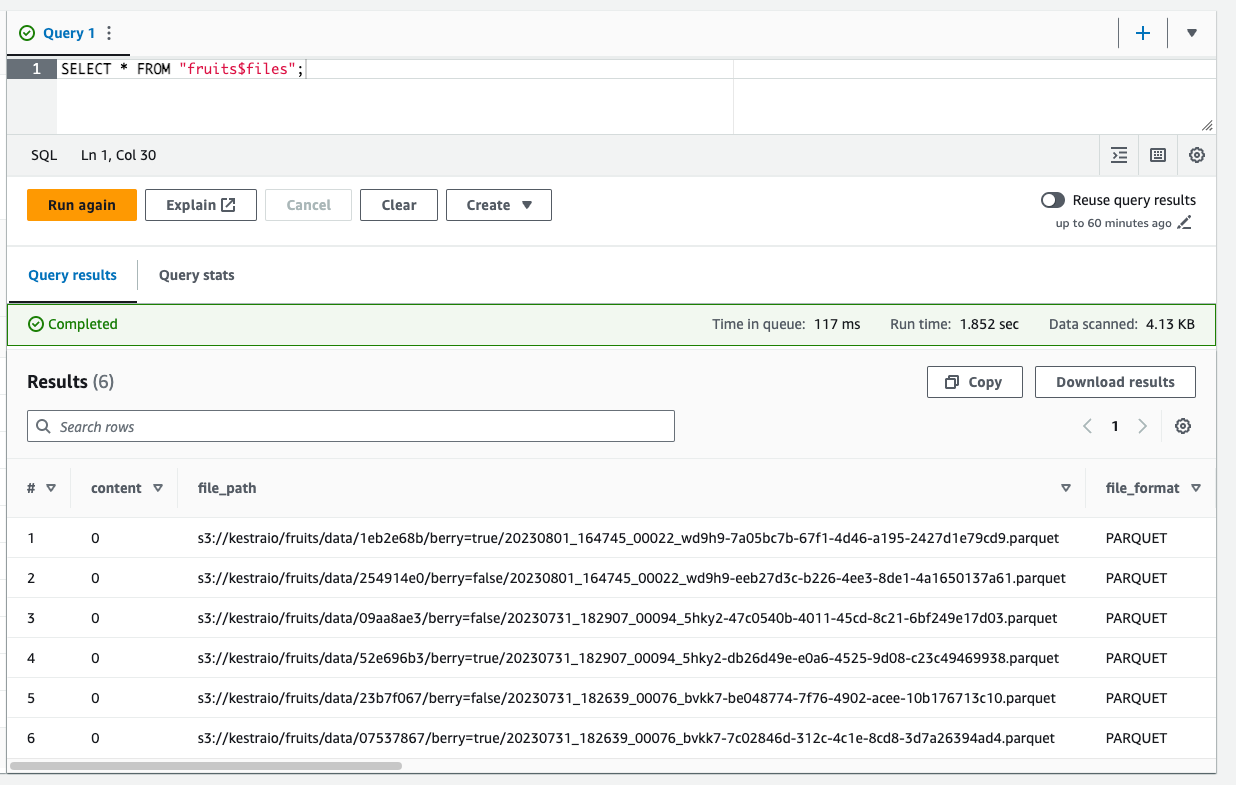
That’s quite a lot of files for such a small dataset. Luckily, Iceberg provides a simple SQL statement to help with that issue.
Time for some Iceberg magic: OPTIMIZE 🪄
The common challenge in managing data lakes is the “Small Files Problem”, where a large number of small files leads to wasted storage, slower reads and longer processing times. Iceberg provides a simple solution to that problem — the OPTIMIZE operation.
The OPTIMIZE table REWRITE DATA is a SQL statement in Amazon Athena that, under the hood, uses Iceberg stored procedures to consolidate small files into bigger files optimized for analytics. The command shown below will automatically rewrite data stored in S3 based on their size, partitioning schema and the number of associated delete files. Magic! 🪄
OPTIMIZE fruits REWRITE DATA USING BIN_PACK;Let’s look at the files again:
SELECT record_count, file_pathFROM "fruits$files";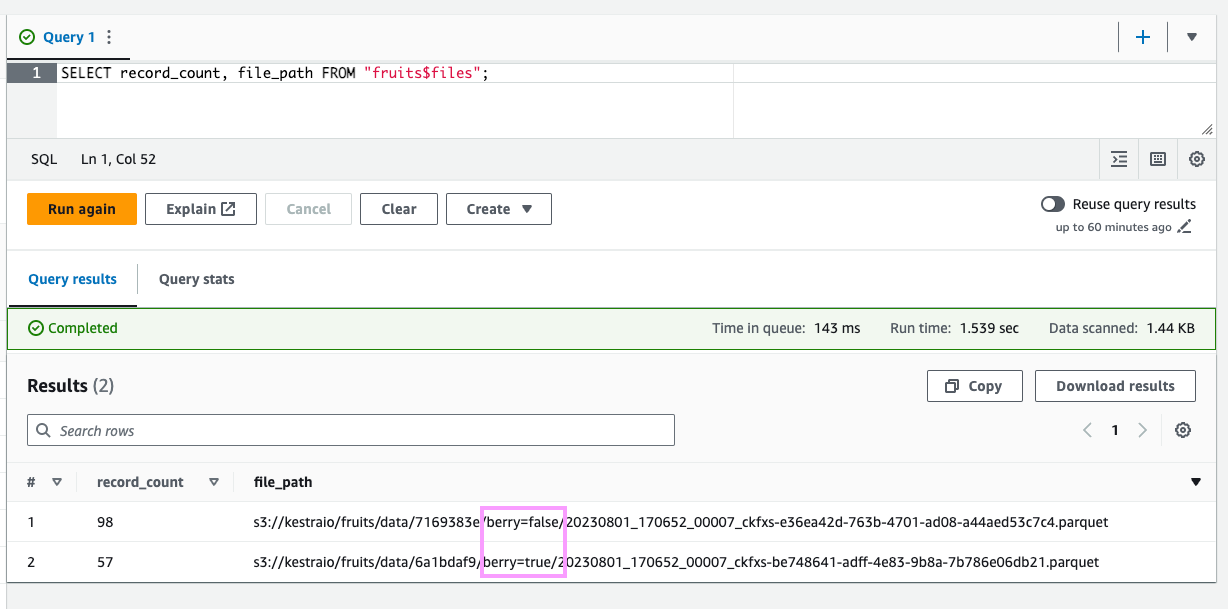
Iceberg automatically clustered all berries and non-berries together into separate files based on the partitioning scheme we provided when creating the table.
Iceberg magic part 2: VACUUM 🪄
Iceberg provides even more useful operations for data management. The VACUUM statement will do the following:
- expire snapshots i.e. the files that exceeded the retention period specified for the table
- remove orphan files i.e. files no longer referenced in any metadata files.
VACUUM fruits;Again, a very powerful operation that can save you hours of hard work.
Ingesting new data into the data lake on a regular basis
We’ve covered a lot of ground so far. We created an Iceberg table, inserted new rows, updated and deleted existing rows, and optimized the table to improve performance. There is one remaining aspect: our data lake should stay up to date. We can either ingest new data on a regular cadence using scheduled batch pipelines or use an event-driven approach to update our Iceberg tables when new files arrive in S3. Let’s look at both patterns.
Python script to ingest new data
First, let’s create a Python script that will ingest new data into our table. You can download the CSV files that start with fruit from the kestra-io/datasets repository and add them to the same directory as your Python script.
We’ll use the same code as before with the following adjustments:
- The
wr.catalog.delete_table_if_exists(...)method will delete the temporary table if it exists. This allows us to make the pipeline idempotent, i.e. running this script multiple times will result in the same state in the data lake without side effects. - When orchestrating this process from Kestra, the line
Kestra.counter("nr_rows", nr_rows, {"table": TABLE})will send a metric to Kestra’s backend to track the number of rows ingested into the data lake over time. This is useful for monitoring and troubleshooting. Running this script locally won’t send anything to Kestra’s backend — it will only print the metric to thestdout. - Remember that we don’t want blueberries or bananas 🍌 in the final dataset? We’ll remove them as part of a data cleaning step in Pandas.
import sysimport awswrangler as wrfrom kestra import Kestra
INGEST_S3_KEY_PATH = "s3://kestraio/inbox/"
if len(sys.argv) > 1: INGEST_S3_KEY_PATH = sys.argv[1]else: print(f"No custom path provided. Using the default path: {INGEST_S3_KEY_PATH}.")
# Iceberg tableBUCKET_NAME = "kestraio"DATABASE = "default"TABLE = "raw_fruits"
# Iceberg table's locationS3_PATH = f"s3://{BUCKET_NAME}/{TABLE}"S3_PATH_TMP = f"{S3_PATH}_tmp"
MERGE_QUERY = """MERGE INTO fruits f USING raw_fruits r ON f.fruit = r.fruit WHEN MATCHED THEN UPDATE SET id = r.id, berry = r.berry, update_timestamp = current_timestamp WHEN NOT MATCHED THEN INSERT (id, fruit, berry, update_timestamp) VALUES(r.id, r.fruit, r.berry, current_timestamp);"""
if not INGEST_S3_KEY_PATH.startswith("s3://"): INGEST_S3_KEY_PATH = f"s3://{BUCKET_NAME}/{INGEST_S3_KEY_PATH}"
df = wr.s3.read_csv(INGEST_S3_KEY_PATH)nr_rows = df.id.nunique()print(f"Ingesting {nr_rows} rows")Kestra.counter("nr_rows", nr_rows, {"table": TABLE})
df = df[~df["fruit"].isin(["Blueberry", "Banana"])]df = df.drop_duplicates(subset=["fruit"], ignore_index=True, keep="first")
wr.catalog.delete_table_if_exists(database=DATABASE, table=TABLE)
wr.athena.to_iceberg( df=df, database=DATABASE, table=TABLE, table_location=S3_PATH, temp_path=S3_PATH_TMP, partition_cols=["berry"], keep_files=False,)
wr.athena.start_query_execution( sql=MERGE_QUERY, database=DATABASE, wait=True,)print(f"New data successfully ingested into {S3_PATH}")As long as you installed the required libraries, you can run this script from a local environment. Next, we’ll see how to orchestrate this process using Kestra.
Scheduled data pipeline
We can create a scheduled data pipeline that will check for new files in S3 every hour. The flow shown below will:
- Check if there are any new files under a given S3 prefix
- Download new CSV files from S3 (if available)
- Read those CSV files as a dataframe, use Pandas to clean the data and ingest it into the S3 data lake managed by Iceberg and AWS Glue
- Move the already ingested file to a different folder to avoid duplicated computations.
id: ingestToDataLakeInlinePythonnamespace: company.team
variables: bucket: kestraio prefix: inbox database: default
tasks: - id: listObjects type: io.kestra.plugin.aws.s3.List prefix: "{{vars.prefix}}" accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" bucket: "{{vars.bucket}}"
- id: check type: io.kestra.plugin.core.flow.If condition: "{{outputs.listObjects.objects}}" then: - id: ingestToDataLake type: io.kestra.plugin.scripts.python.Script warningOnStdErr: false env: AWS_ACCESS_KEY_ID: "{{ secret('AWS_ACCESS_KEY_ID') }}" AWS_SECRET_ACCESS_KEY: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" AWS_DEFAULT_REGION: "{{ secret('AWS_DEFAULT_REGION') }}" docker: image: ghcr.io/kestra-io/aws:latest script: | import awswrangler as wr from kestra import Kestra
# Iceberg table BUCKET_NAME = "{{vars.bucket}}" DATABASE = "{{vars.database}}" TABLE = "raw_fruits"
# Iceberg table's location S3_PATH = f"s3://{BUCKET_NAME}/{TABLE}" S3_PATH_TMP = f"{S3_PATH}_tmp"
# File to ingest PREFIX = "{{vars.prefix}}" INGEST_S3_KEY_PATH = f"s3://{BUCKET_NAME}/{PREFIX}/"
df = wr.s3.read_csv(INGEST_S3_KEY_PATH) nr_rows = df.id.nunique() print(f"Ingesting {nr_rows} rows") Kestra.counter("nr_rows", nr_rows, {"table": TABLE})
df = df[~df["fruit"].isin(["Blueberry", "Banana"])] df = df.drop_duplicates(subset=["fruit"], ignore_index=True, keep="first")
wr.catalog.delete_table_if_exists(database=DATABASE, table=TABLE)
wr.athena.to_iceberg( df=df, database=DATABASE, table=TABLE, table_location=S3_PATH, temp_path=S3_PATH_TMP, partition_cols=["berry"], keep_files=False, ) print(f"New data successfully ingested into {S3_PATH}")
- id: mergeQuery type: io.kestra.plugin.aws.athena.Query accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" database: "{{vars.database}}" outputLocation: "s3://{{vars.bucket}}/query_results/" query: | MERGE INTO fruits f USING raw_fruits r ON f.fruit = r.fruit WHEN MATCHED THEN UPDATE SET id = r.id, berry = r.berry, update_timestamp = current_timestamp WHEN NOT MATCHED THEN INSERT (id, fruit, berry, update_timestamp) VALUES(r.id, r.fruit, r.berry, current_timestamp);
- id: optimize type: io.kestra.plugin.aws.athena.Query accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" database: "{{vars.database}}" outputLocation: "s3://{{vars.bucket}}/query_results/" query: | OPTIMIZE fruits REWRITE DATA USING BIN_PACK;
- id: moveToArchive type: io.kestra.plugin.aws.cli.AwsCLI accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" commands: - aws s3 mv s3://{{vars.bucket}}/{{vars.prefix}}/ s3://{{vars.bucket}}/archive/{{vars.prefix}}/ --recursive
triggers: - id: hourlySchedule type: io.kestra.plugin.core.trigger.Schedule cron: "@hourly" disabled: trueWhen you execute the flow, you should see the following output in the logs:
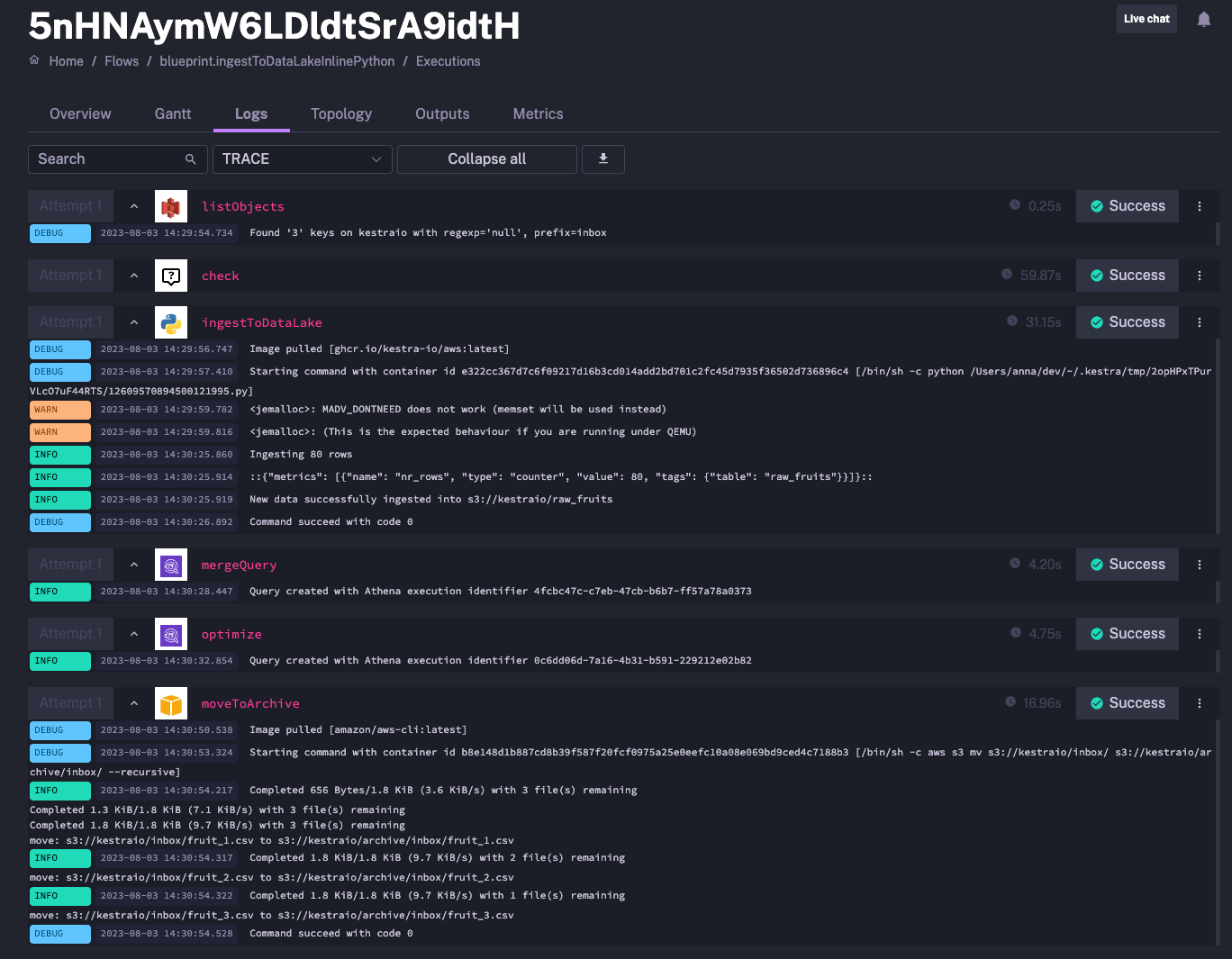
Once you push your script to Git, your pipeline can be simplified as follows:
id: ingestToDataLakeGitnamespace: company.team
variables: bucket: kestraio prefix: inbox database: default
tasks: - id: listObjects type: io.kestra.plugin.aws.s3.List prefix: "{{vars.prefix}}" accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" bucket: "{{vars.bucket}}"
- id: check type: io.kestra.plugin.core.flow.If condition: "{{outputs.listObjects.objects}}" then: - id: processNewObjects type: io.kestra.plugin.core.flow.WorkingDirectory tasks: - id: git type: io.kestra.plugin.git.Clone url: https://github.com/kestra-io/scripts branch: main
- id: ingestToDataLake type: io.kestra.plugin.scripts.python.Commands warningOnStdErr: false env: AWS_ACCESS_KEY_ID: "{{ secret('AWS_ACCESS_KEY_ID') }}" AWS_SECRET_ACCESS_KEY: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" AWS_DEFAULT_REGION: "{{ secret('AWS_DEFAULT_REGION') }}" docker: image: ghcr.io/kestra-io/aws:latest commands: - python etl/aws_iceberg_fruit.py
- id: mergeQuery type: io.kestra.plugin.aws.athena.Query accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" database: "{{vars.database}}" outputLocation: "s3://{{vars.bucket}}/query_results/" query: | MERGE INTO fruits f USING raw_fruits r ON f.fruit = r.fruit WHEN MATCHED THEN UPDATE SET id = r.id, berry = r.berry, update_timestamp = current_timestamp WHEN NOT MATCHED THEN INSERT (id, fruit, berry, update_timestamp) VALUES(r.id, r.fruit, r.berry, current_timestamp);
- id: optimize type: io.kestra.plugin.aws.athena.Query accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" database: "{{vars.database}}" outputLocation: "s3://{{vars.bucket}}/query_results/" query: | OPTIMIZE fruits REWRITE DATA USING BIN_PACK;
- id: moveToArchive type: io.kestra.plugin.aws.cli.AwsCLI accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" commands: - aws s3 mv s3://{{vars.bucket}}/{{vars.prefix}}/ s3://{{vars.bucket}}/archive/{{vars.prefix}}/ --recursive
triggers: - id: hourlySchedule type: io.kestra.plugin.core.trigger.Schedule cron: "@hourly"The kestra-io/scripts is a public repository, so you can reproduce that flow directly as long as you adjust the S3 bucket name and set your AWS credentials. To leverage private repositories, you only need to add your Git user name and a Personal Access Token. Check the Secrets docs to see how to manage sensitive values in Kestra.
Executing this workflow should give you the following output:
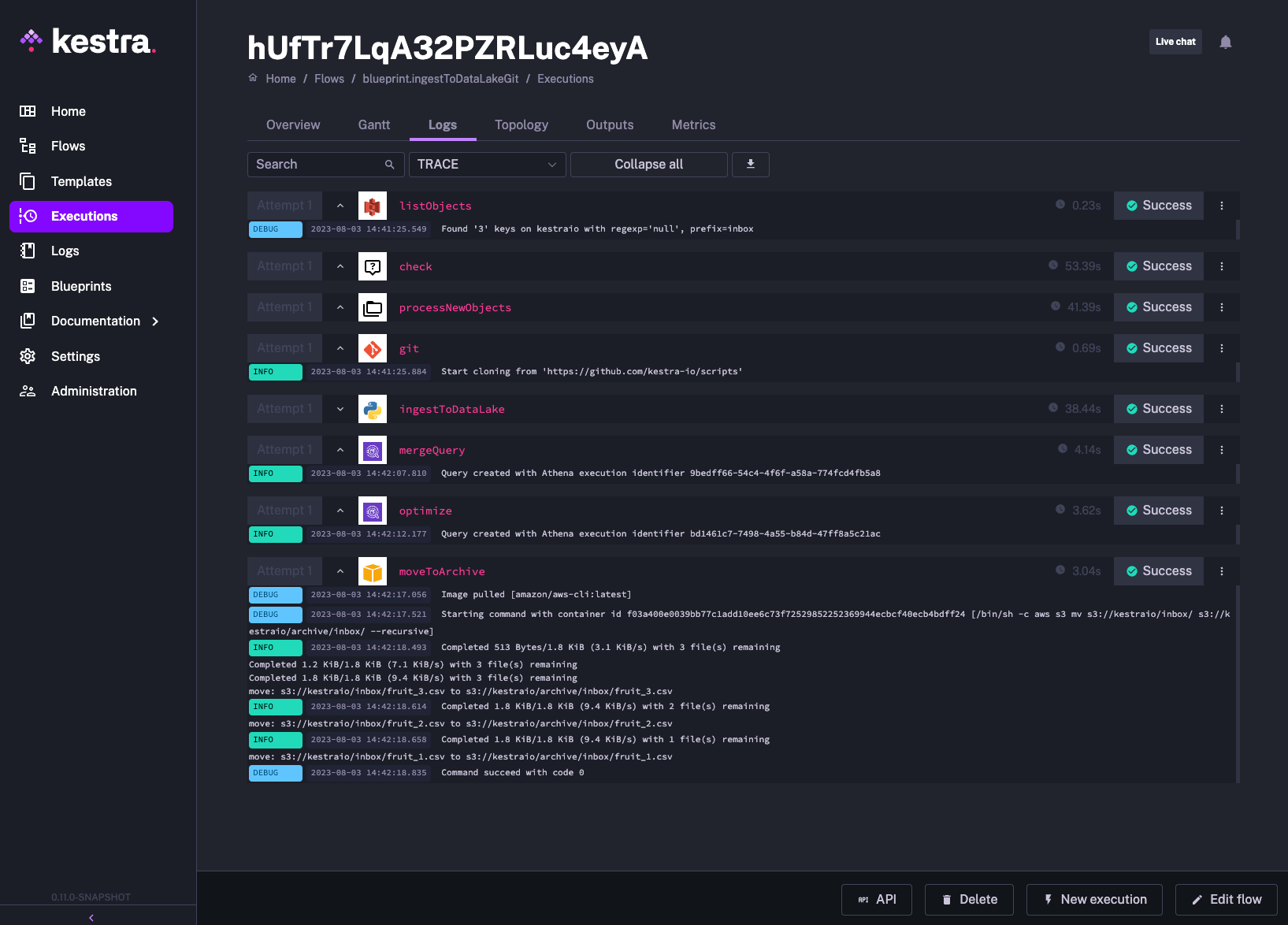
This scheduled workflow is simple to understand and easy to run locally. However, in certain circumstances, it might be inefficient. External systems are often unpredictable, making it difficult to figure out the optimal batch pipeline frequency. The flow shown above will run every hour, even if there are no new files in S3 for days or weeks. In such scenarios, event triggers and a decoupled approach to pipelines become incredibly useful.
Event-driven data pipeline
Kestra makes it easy to switch between scheduled and event-driven workflows simply by adjusting the trigger configuration.
The flow code below uses the same Python script we used for a scheduled workflow. The only difference is that, when calling the script, we now pass the detected S3 object key from the trigger as an input argument. Then, the script transforms and loads data to the S3 data lake exactly the same way as before.
You can see here a significant advantage of Kestra: a separation of concerns between orchestration and business logic. You don’t have to modify your code in any way - Kestra can orchestrate your custom code written in any language with no modifications.
id: ingestToDataLakeEventDrivennamespace: company.team
variables: sourcePrefix: inbox destinationPrefix: archive database: default bucket: kestraio
tasks: - id: wdir type: io.kestra.plugin.core.flow.WorkingDirectory tasks: - id: git type: io.kestra.plugin.git.Clone url: https://github.com/kestra-io/scripts
- id: etl type: io.kestra.plugin.scripts.python.Commands warningOnStdErr: false docker: image: ghcr.io/kestra-io/aws:latest env: AWS_ACCESS_KEY_ID: "{{ secret('AWS_ACCESS_KEY_ID') }}" AWS_SECRET_ACCESS_KEY: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" AWS_DEFAULT_REGION: "{{ secret('AWS_DEFAULT_REGION') }}" commands: - python etl/aws_iceberg_fruit.py {{vars.destinationPrefix}}/{{ trigger.objects | jq('.[].key') | first }}
- id: mergeQuery type: io.kestra.plugin.aws.athena.Query accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" database: "{{vars.database}}" outputLocation: "s3://{{vars.bucket}}/query_results/" query: | MERGE INTO fruits f USING raw_fruits r ON f.fruit = r.fruit WHEN MATCHED THEN UPDATE SET id = r.id, berry = r.berry, update_timestamp = current_timestamp WHEN NOT MATCHED THEN INSERT (id, fruit, berry, update_timestamp) VALUES(r.id, r.fruit, r.berry, current_timestamp);
- id: optimize type: io.kestra.plugin.aws.athena.Query accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}" region: "{{ secret('AWS_DEFAULT_REGION') }}" database: "{{vars.database}}" outputLocation: "s3://{{vars.bucket}}/query_results/" query: | OPTIMIZE fruits REWRITE DATA USING BIN_PACK;
triggers: - id: waitForNewS3objects type: io.kestra.plugin.aws.s3.Trigger bucket: kestraio interval: PT1S maxKeys: 1 filter: FILES action: MOVE prefix: "{{vars.sourcePrefix}}" # e.g. s3://kestraio/inbox/fruit_1.csv moveTo: key: "{{vars.destinationPrefix}}/{{vars.sourcePrefix}}" # e.g. s3://kestraio/archive/inbox/fruit_1.csv region: "{{ secret('AWS_DEFAULT_REGION') }}" accessKeyId: "{{ secret('AWS_ACCESS_KEY_ID') }}" secretKeyId: "{{ secret('AWS_SECRET_ACCESS_KEY') }}"The trigger polls the S3 location, indicated by prefix, for new files every second. The flow gets executed as soon as a new S3 key is detected.
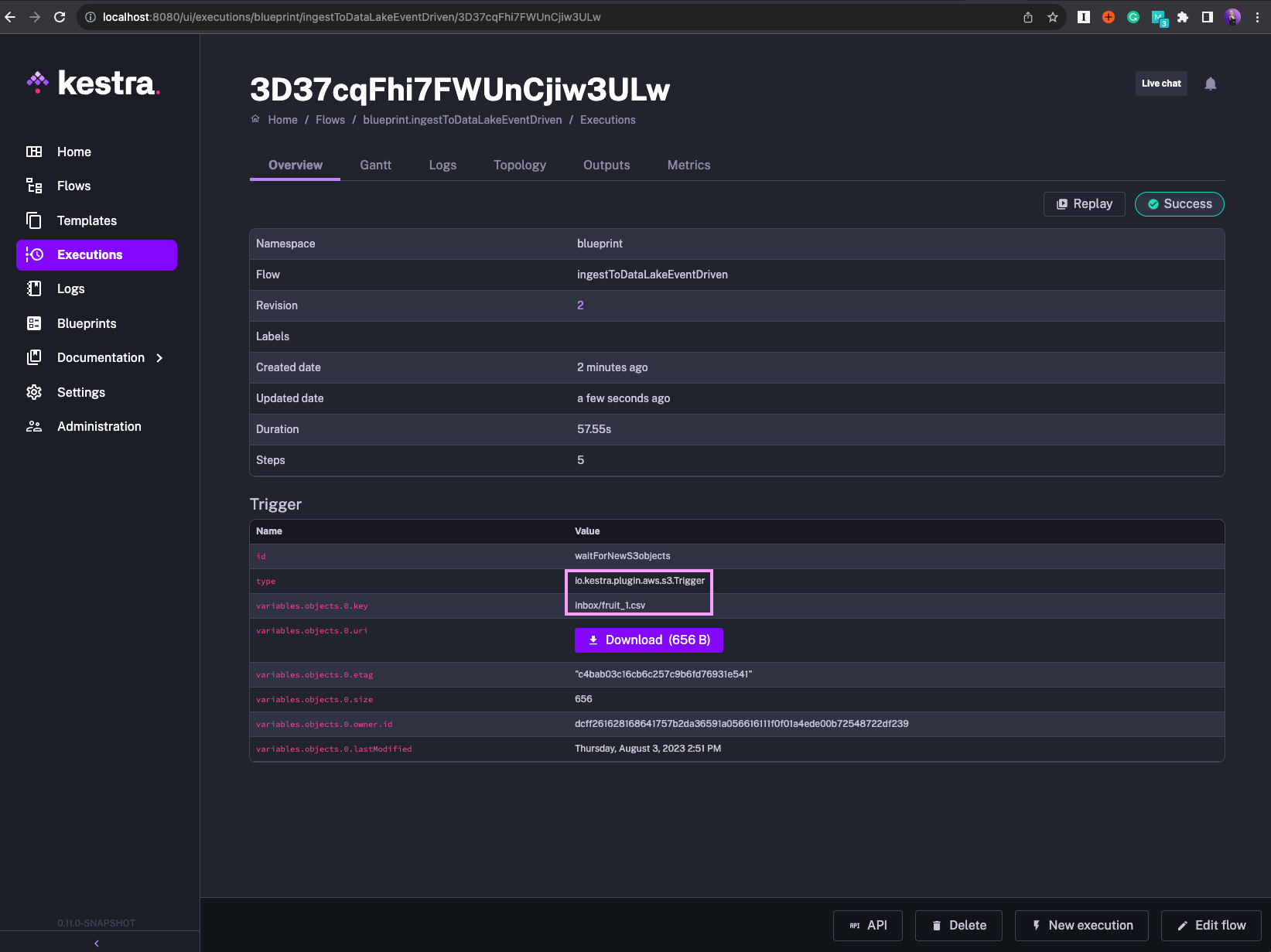
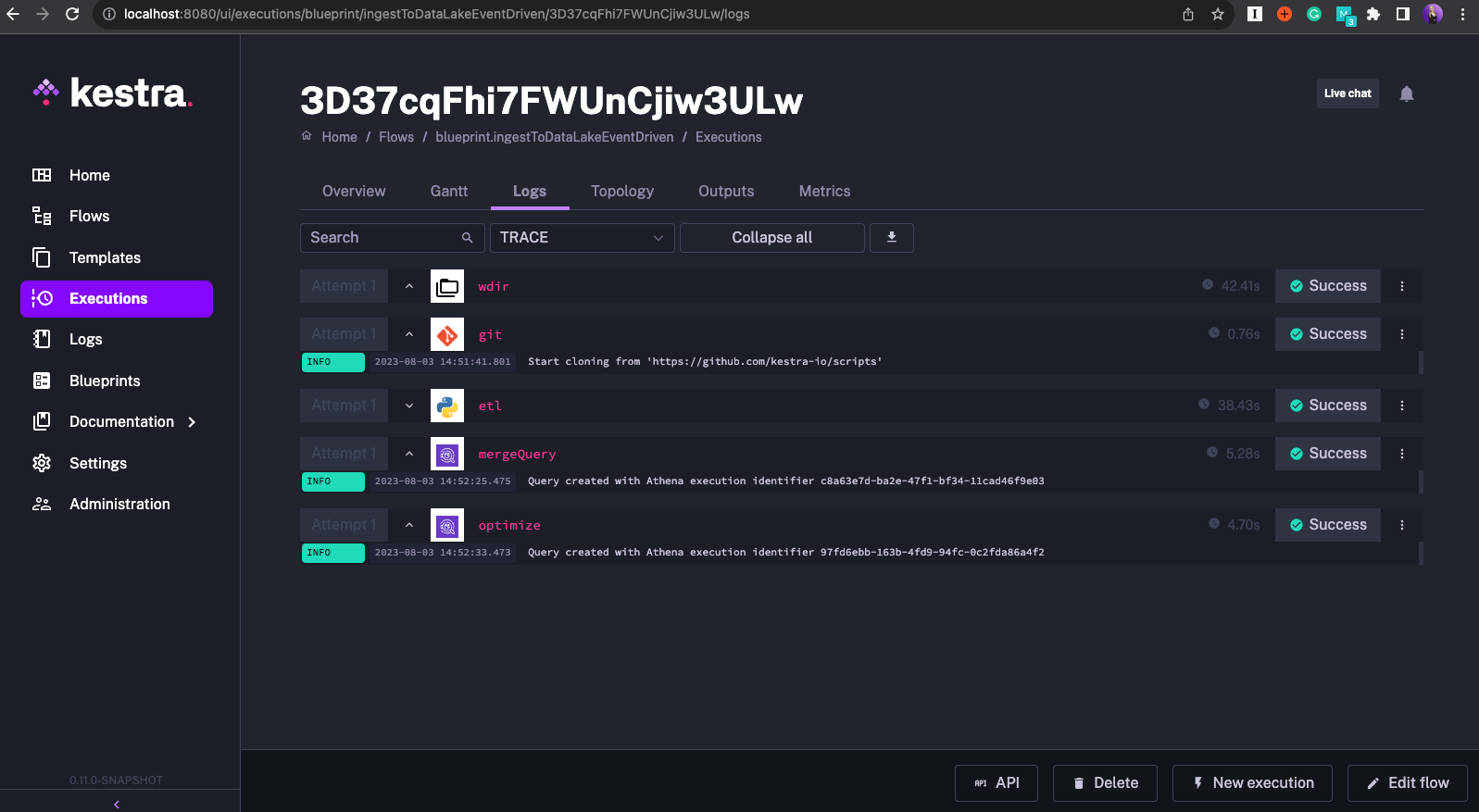
The processed file is moved to the archive directory to avoid the flow being triggered multiple times for the same file. The maxKeys property is particularly helpful in our use case as it allows us to process each incoming file sequentially as if you were using a FIFO queue.
See the screenshots below which demonstrate how the flow was triggered for each new S3 object with the inbox prefix.
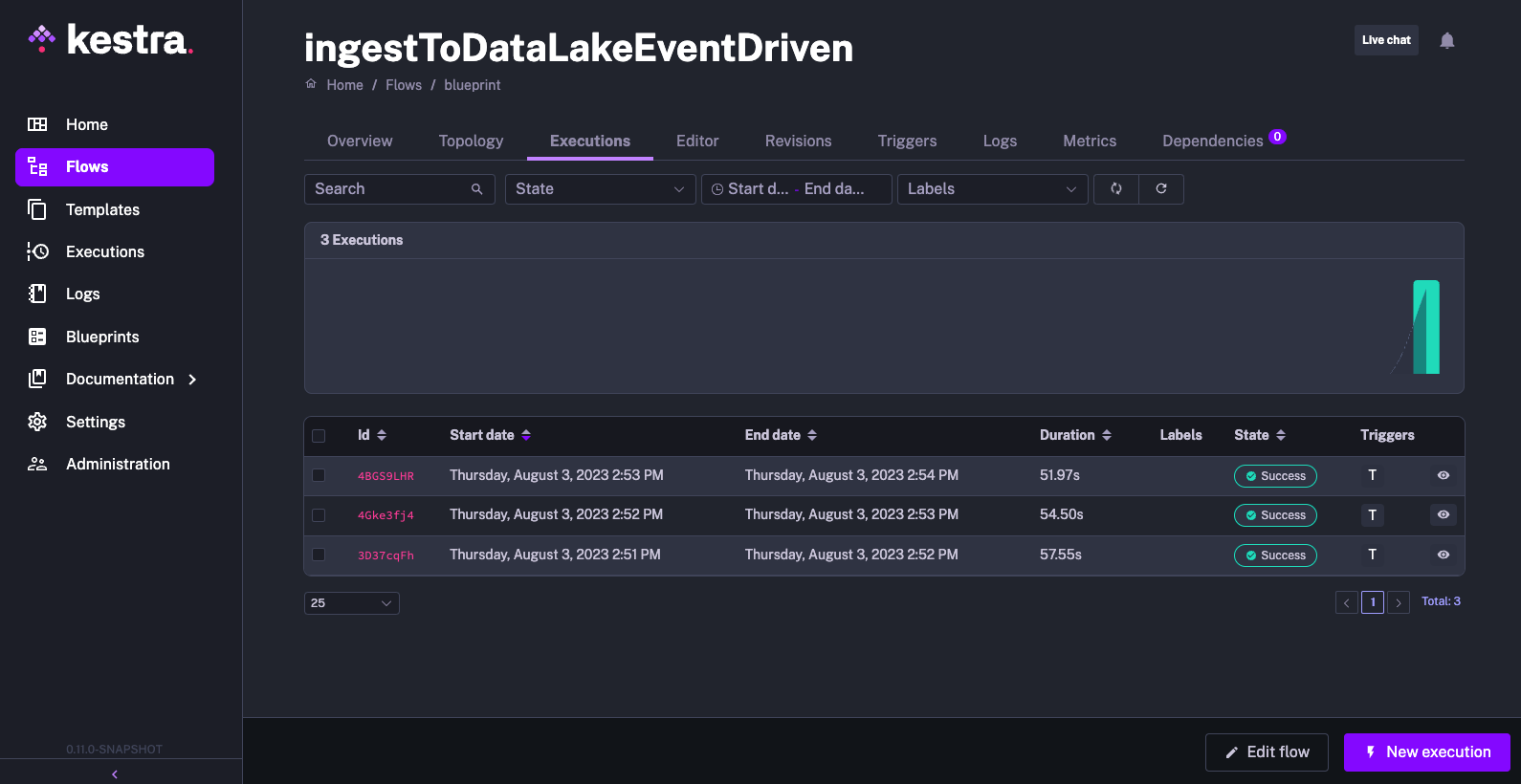
Iceberg Blueprints
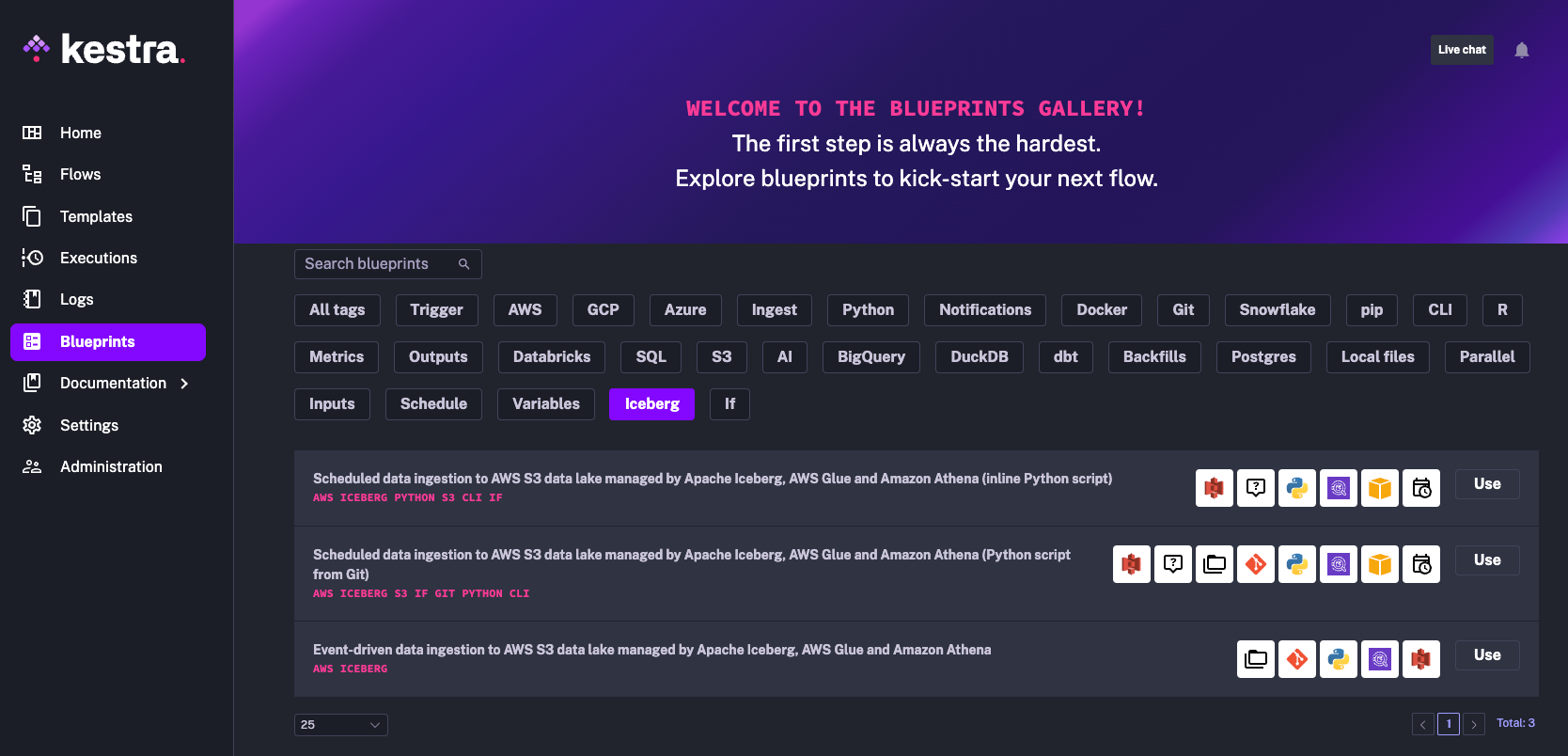
All Kestra workflows covered in this post are available as Blueprints directly from the Kestra UI. Start Kestra using Docker Compose, as explained in the Getting Started guide, and select the Iceberg tag from the Blueprints section:
Next steps
This tutorial demonstrated how to use Apache Iceberg with Amazon Athena, AWS Glue and Amazon S3, and how to manage a scheduled and event-driven data ingestion process with Kestra.
If you have any questions about what we’ve covered in this post, reach out via our community Slack. Lastly, if you like the project, give us a star on GitHub.
Get Kestra updates to your inbox
Stay up to date with the latest features and changes to Kestra